Through a combination of light and electron microscopy techniques, I seek to understand and quantify the biomolecular bases of disease. These methods can create high-fidelity 3-dimensional reconstructions of biological samples across several size scales, from tissues to the cell and its constituent parts, all the way down to individual proteins. These unique insights generate new avenues for therapeutic intervention to combat drug resistance and increase therapeutic efficacy.
Liquid phase electron microscopy (LPEM)
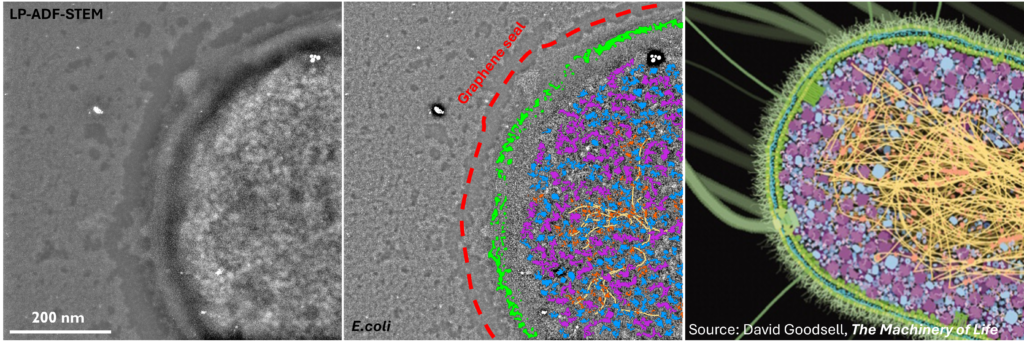
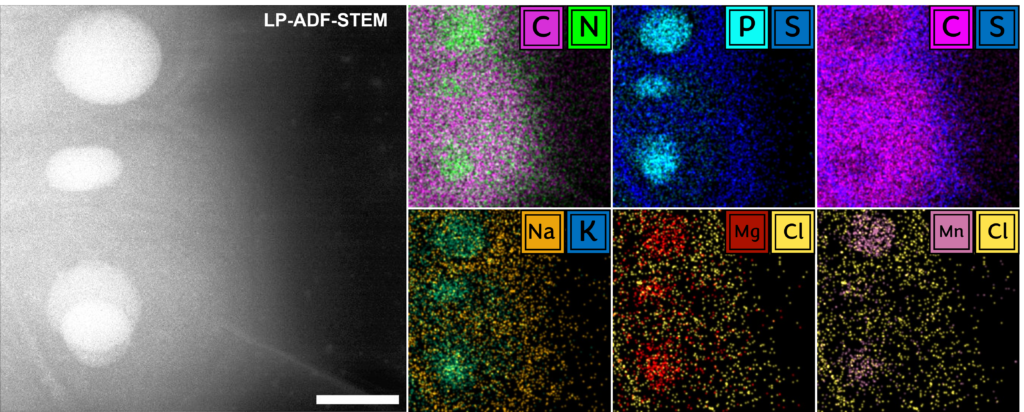
Fundamentally, biology is driven by transient, dynamic assemblies of biomolecules in solution. This method enables the study of structural and dynamic features in truly native environments. Using this technique it may be possible in the near future to link protein structure with cellular function by mapping the dynamic landscape of these intermolecular interactions within living systems. The insights generated from this study will illuminate our understanding of fundamental cellular processes and associated diseases.
cryogenic-electron microscopy (cryo-em)
Cryo-EM analysis allows us to observe proteins in near-native conditions. This method provides us with a near-atomic resolution map (~0.25 nm) of a protein, revealing the fundamental interactions which regulate the structural changes responsible for its activity and malfunction in disease. We can also use this technique to examine the direct effects of small-molecule therapeutics, co-factors and other biomolecules of interest on the conformational landscape of the protein and use this knowledge to inform more effective therapies.
correlative light-electron microscopy (clem)
CLEM techniques combine both light and electron microscopies, complementing each other by merging the large sampling volume and broad selection of fluorescent labels and applications (e.g. Förster Resonance Energy Transfer (FRET), Fluorescence Lifetime Imaging (FLIM) etc…) in light microscopy with the extremely high resolving power of the electron microscope.
focused ion beam - scanning electron microscopy (fib-sem)
We developed a high-throughput method of acquiring and segmenting large volumes of human tissues, demonstrating that large-scale quantitative statistical analyses at nanometre-level detail in human tissues is possible with FIB-SEM methods. Over 1 million μm3 across 10 individuals at 15 nm3 pixel resolution in XYZ acquired and segmented using our approach, representing a 500x increase in throughput relative to conventional methods. This technique allows us to explore the ultrastructural causes of disease at an unprecedented scale and at an extremely high level of detail.
latest publications
Liquid Phase EM
, B.J., , A., , E., , B., , J.S. and “Liquid phase electron microscopy of bacterial ultrastructure” Small, 20 (50), 2402871.
Cryo-EM
Caffrey, B. J., Zhu, X., Berezuk, A., Tuttle, K., Chittori, S. and Subramaniam, S. (2021) “Cryo-EM reveals disrupted human p97 allosteric activation by disease mutations and inhibitor binding” Journal of Biological Chemistry 297 (4), 101187.
CLEM
Caffrey, B. J. (2021) “Examining Nanoparticle Interactions on a Cellular Scale Using Correlative Light Electron Microscopy.” Molecular Biology and Nanomedicine 2(1), 20–27.
FIB-SEM
Caffrey, B. J., Maltsev, A. V., Gonzalez-Freire, M., Hartnell, L. M., Ferrucci, L. and Subramaniam, S. (2019) “Semi-automated 3D segmentation of human skeletal muscle using Focused Ion Beam-Scanning Electron Microscopic images.” Journal of Structural Biology 207 (1), 1–11.
Literature Review
Caffrey, B. J. and Subramaniam, S. (2022) “Imaging Cellular Architecture in Three Dimensions Through Electron Microscopy.” In Bradshaw, R., Stahl, P. & Hart, J. (eds.) The Encyclopedia of Cell Biology 2nd ed.
contact